
时间:2024-03-05 22:27:46 阅读:142

bilibili:文章对应的讲解视频在此。熊大学习社 https://space.bilibili.com/475774512
Gitee开源:https://gitee.com/shenghuaxiong/ioter
CSDN玩转物联网专栏文章:https://blog.csdn.net/shx13141/category_11669532.html
微信公众号:熊大学习社
课程资料包括糖尿病预测模型代码(SVM、随机森林、逻辑回归、K近邻、梯度提升模型,基于python)、论文的NHANES数据、复现论文、讲义。关注公众号“熊大学习社”,回复“t221201”,可获得资料链接。谢谢您的支持,我们坚持学以致用、高效学习、质量服务,做好有质量的分享。
另外,我们提供专业的SCI论文指导,服务合作请加vx:learning-daxiong
论文:An Ensemble Classifier for Predicting the Onset of Type II Diabetes

期刊:arXiv stat.ML,计算机机器学习

安装python和jupyter notebook的参考方法,在B站搜索即可,如下。`
【一劳永逸安装和配置python3.7.2(新手必读)】 https://www.bilibili.com/video/BV18b411q7WN/?share_source=copy_web&vd_source=ffe42a784613eb42f19e6b01511f4cd8
【python数据分析神器Jupyter notebook快速入门】 https://www.bilibili.com/video/BV1Q4411H7fJ/?share_source=copy_web&vd_source=ffe42a784613eb42f19e6b01511f4cd8
代码在dataprepared.ipynb
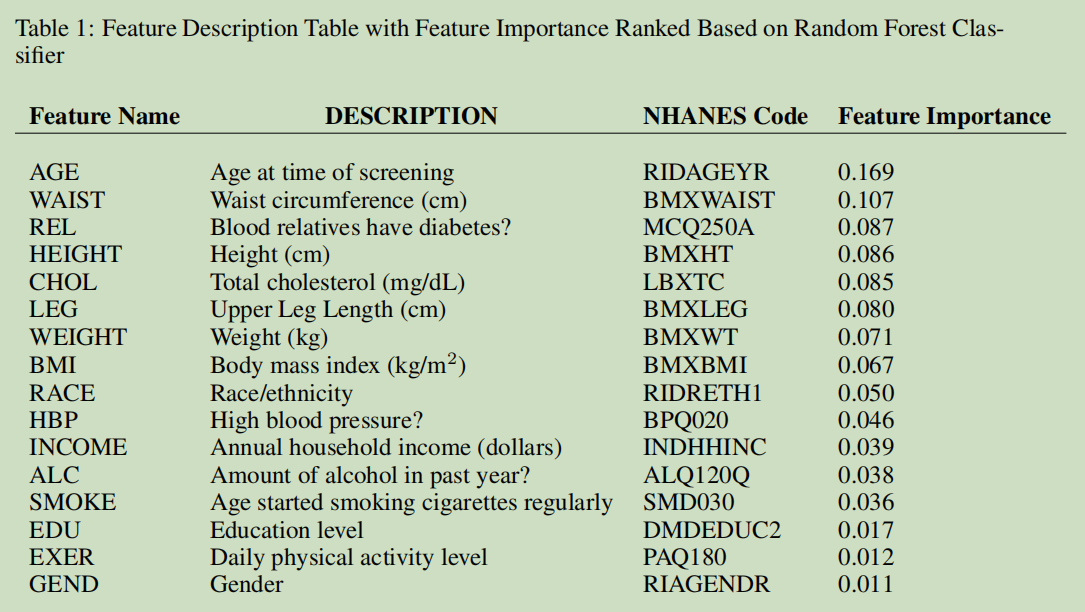
自变量+因变量,这是自变量。
ALQ120Q:
BMXBMI:
BMXHT:
BMXLEG:
BMXWAIST:
BMXWT:
BPQ020:
DMDEDUC2:
INDHHINC:
LBXTC:
MCQ250A:
PAQ180:
RIAGENDR:
RIDAGEYR:
RIDRETH1:
SMD030:
数据准备:
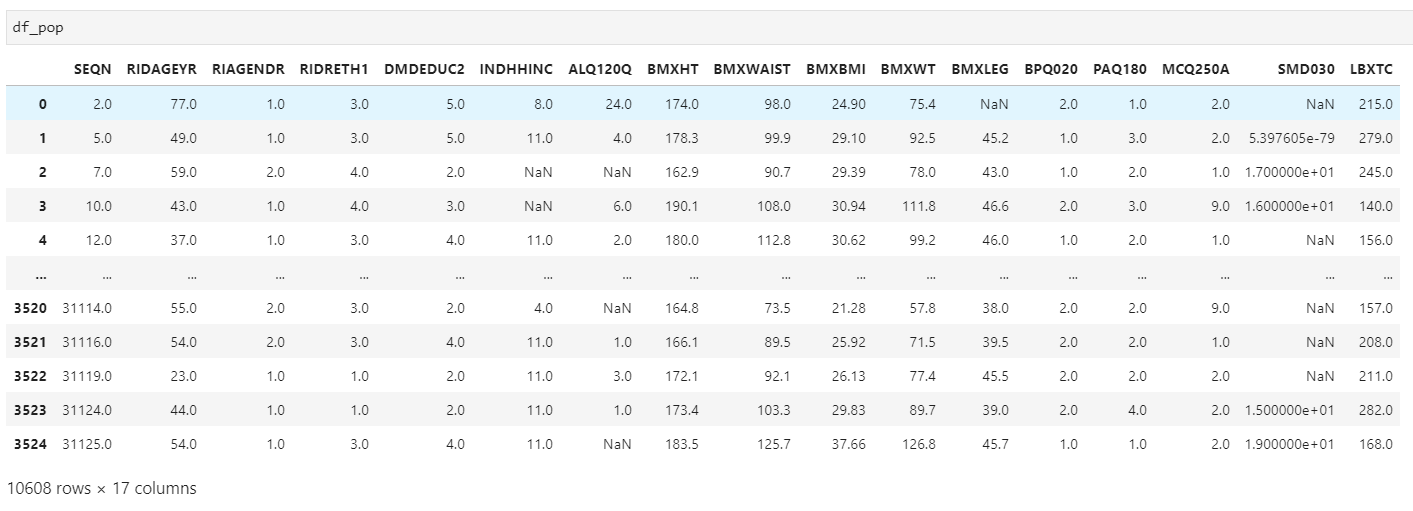
# 数据整合
'''Merge Datasets
Age >= 20 and non-pregnant population
'''
logging.info('Merging data...')
age = 20
# 1999-2000
df_00 = DEMO.loc[((DEMO.RIAGENDR == 2) & (DEMO.RIDAGEYR >= age) & (DEMO.RIDEXPRG == 2)) |
((DEMO.RIAGENDR == 1) & (DEMO.RIDAGEYR >= age)),
DEMO_cols]
.merge(ALQ[ALQ_cols], on='SEQN')
.merge(BMX[BMX_cols], on='SEQN')
.merge(BPQ[BPQ_cols], on='SEQN')
.merge(PAQ[PAQ_cols], on='SEQN')
.merge(MCQ[MCQ_cols], on='SEQN')
.merge(SMQ[SMQ_cols], on='SEQN')
.merge(LAB13[LAB13_cols], on='SEQN')
# 2001-2002
df_02 = DEMO_B.loc[((DEMO_B.RIAGENDR == 2) & (DEMO_B.RIDAGEYR >= age) & (DEMO_B.RIDEXPRG == 2)) |
((DEMO_B.RIAGENDR == 1) & (DEMO_B.RIDAGEYR >= age)),
DEMO_cols]
.merge(ALQ_B[ALQ_cols], on='SEQN')
.merge(BMX_B[BMX_cols], on='SEQN')
.merge(BPQ_B[BPQ_cols], on='SEQN')
.merge(PAQ_B[PAQ_cols], on='SEQN')
.merge(MCQ_B[MCQ_cols], on='SEQN')
.merge(SMQ_B[SMQ_cols], on='SEQN')
.merge(LAB13_B[LAB13_cols], on='SEQN')
# 2003-2004
df_04 = DEMO_C.loc[((DEMO_C.RIAGENDR == 2) & (DEMO_C.RIDAGEYR >= age) & (DEMO_C.RIDEXPRG == 2)) |
((DEMO_C.RIAGENDR == 1) & (DEMO_C.RIDAGEYR >= age)),
DEMO_cols]
.merge(ALQ_C[ALQ_cols], on='SEQN')
.merge(BMX_C[BMX_cols], on='SEQN')
.merge(BPQ_C[BPQ_cols], on='SEQN')
.merge(PAQ_C[PAQ_cols], on='SEQN')
.merge(MCQ_C[MCQ_cols], on='SEQN')
.merge(SMQ_C[SMQ_cols], on='SEQN')
.merge(LAB13_C[LAB13_cols], on='SEQN')
# 不同年份组合
df_pop = pd.concat([df_00, df_02, df_04])
代码在dataprepared.ipynb
自变量+因变量,这是因变量。
判断标准:DIQ.DIQ010 == 1
https://wwwn.cdc.gov/Nchs/Nhanes/1999-2000/DIQ.htm#DIQ010
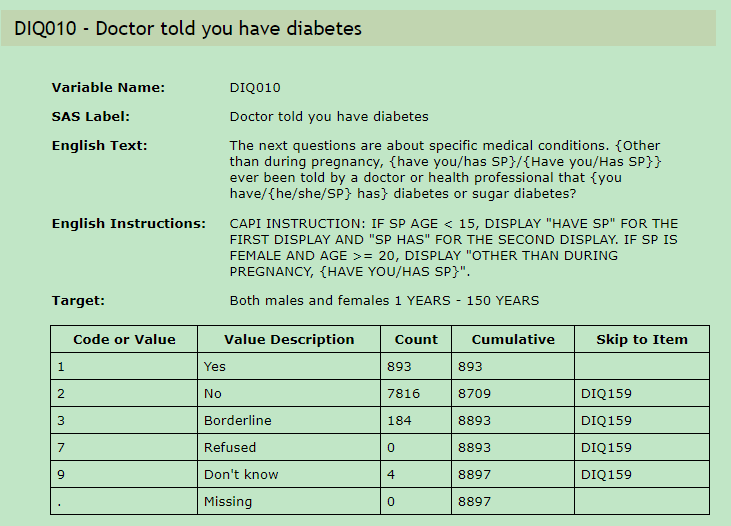
# 确诊的糖尿病患者数据统计
'''Diagnosed Diabetes (total should be 1,266)
The next questions are about specific medical conditions.
{Other than during pregnancy, {have you/has SP}/{Have you/Has SP}}
ever been told by a doctor or health professional that
{you have/{he/she/SP} has} diabetes or sugar diabetes?
'''
df_00_diag = df_00.merge(DIQ.loc[DIQ.DIQ010 == 1, DIQ_cols], on="SEQN")
df_02_diag = df_02.merge(DIQ_B.loc[DIQ_B.DIQ010 == 1, DIQ_cols], on="SEQN")
df_04_diag = df_04.merge(DIQ_C.loc[DIQ_C.DIQ010 == 1, DIQ_cols], on="SEQN")
diag_total = pd.concat([df_00_diag, df_02_diag, df_04_diag])
diag_total.loc[:,'status'] = 1
logging.info('Diagnosed subject count: {}'.format(diag_total.shape[0]))
判定标准:DIQ.DIQ010 == 2,LAB10AM_B.LBXGLU >= 126
https://wwwn.cdc.gov/Nchs/Nhanes/1999-2000/DIQ.htm#DIQ010
https://wwwn.cdc.gov/Nchs/Nhanes/1999-2000/LAB10AM.htm#LBXGLU
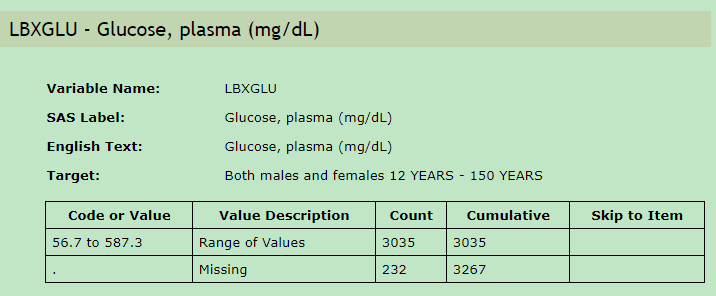
# 未确诊的糖尿病患者数据统计
'''Undiagnosed Diabetes (total should be 195)'''
df_00_undiag = df_00.merge(DIQ.loc[DIQ.DIQ010 == 2, DIQ_cols]
.merge(LAB10AM.loc[LAB10AM.LBXGLU >= 126, LAB10AM_cols], on='SEQN'),
on='SEQN')
df_02_undiag = df_02.merge(DIQ_B.loc[DIQ_B.DIQ010 == 2, DIQ_cols]
.merge(LAB10AM_B.loc[LAB10AM_B.LBXGLU >= 126, LAB10AM_cols], on='SEQN'),
on='SEQN')
df_04_undiag = df_04.merge(DIQ_C.loc[DIQ_C.DIQ010 == 2, DIQ_cols]
.merge(LAB10AM_C.loc[LAB10AM_C.LBXGLU >= 126, LAB10AM_cols], on='SEQN'),
on='SEQN')
undiag_total = pd.concat([df_00_undiag, df_02_undiag, df_04_undiag])
undiag_total.loc[:,'status'] = 1
logging.info('Undiagnosed subject count: {}'.format(undiag_total.shape[0]))
判断标准:(LAB10AM.LBXGLU >= 100) & (LAB10AM.LBXGLU <= 125)
# 糖尿病前期患者数据统计
'''Pre-diabetes (total should be 1,576)'''
df_00_prediab = df_00.merge(LAB10AM.loc[(LAB10AM.LBXGLU >= 100) &
(LAB10AM.LBXGLU <= 125),
LAB10AM_cols], on='SEQN')
df_02_prediab = df_02.merge(LAB10AM_B.loc[(LAB10AM_B.LBXGLU >= 100) &
(LAB10AM_B.LBXGLU <= 125),
LAB10AM_cols], on='SEQN')
df_04_prediab = df_04.merge(LAB10AM_C.loc[(LAB10AM_C.LBXGLU >= 100) &
(LAB10AM_C.LBXGLU <= 125),
LAB10AM_cols], on='SEQN')
prediab_total = pd.concat([df_00_prediab, df_02_prediab, df_04_prediab])
prediab_total.loc[:,'status'] = 0
logging.info('Pre-diabetic subject count: {}'.format(prediab_total.shape[0]))
判定标准:LAB10AM.LBXGLU < 100
# 非糖尿病患者数据统计
'''No Diabetes (total should be 3,277)'''
df_00_nodiab = df_00.merge(LAB10AM.loc[LAB10AM.LBXGLU < 100, LAB10AM_cols], on='SEQN')
df_02_nodiab = df_02.merge(LAB10AM_B.loc[LAB10AM_B.LBXGLU < 100, LAB10AM_cols], on='SEQN')
df_04_nodiab = df_04.merge(LAB10AM_C.loc[LAB10AM_C.LBXGLU < 100, LAB10AM_cols], on='SEQN')
nodiab_total = pd.concat([df_00_nodiab, df_02_nodiab, df_04_nodiab])
nodiab_total.loc[:,'status'] = 0
logging.info('Diabetes-free subject count: {}'.format(nodiab_total.shape[0]))
代码在NHANES.ipynb
5个模型:
Logistic Regression:逻辑回归
KNN:K近邻
Random Forests:随机森林
Gradient Boosting:梯度提升
Support Vector Machine, SVM:支持向量机
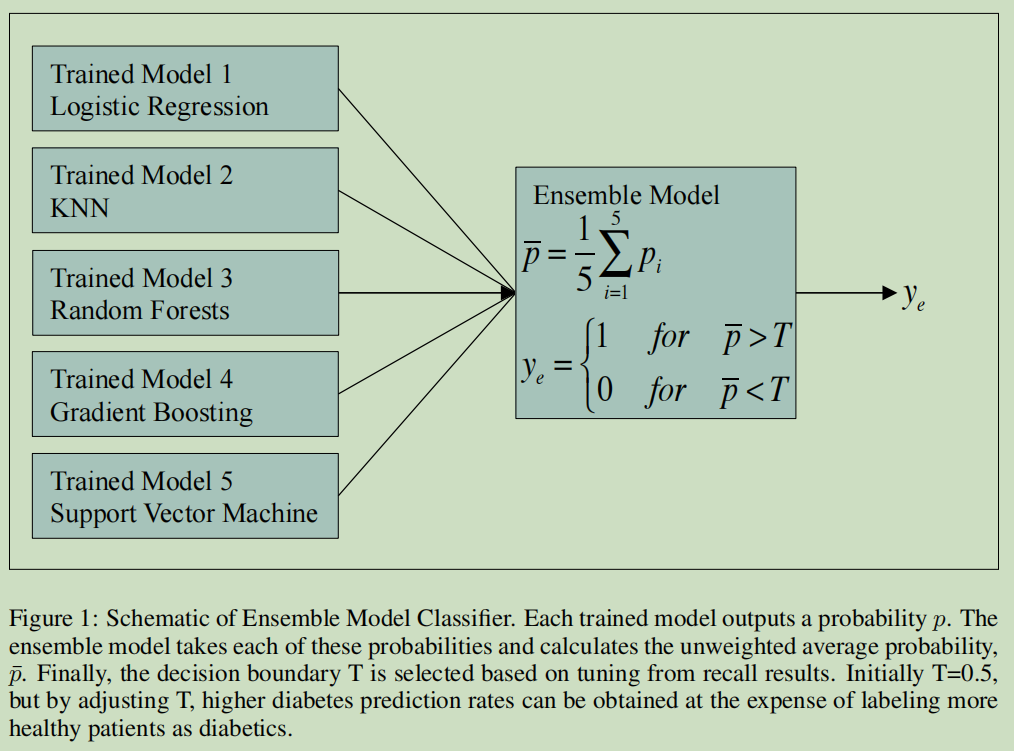
实现过程如下图,具体过程结合代码讲解。

(1)相比于模型的原理,模型怎么用更重要,仅代表个人观点。模型怎么用,再回顾一下,这里最重要的是接口,也就是输入和输出。
(2)站在巨人的肩膀上学习。这篇论文就是这样的,以前做过研究,现在进一步改进。怎么改进:一是再增加2个指标;二是预测算法再增加一些比较。
(3)论文绘图怎么调。实操一下。
(4)算法应用的基本思路:确定模型、模型的评价指标、ROC等。
Precision,Recall,F1score,以及accuracy
https://blog.csdn.net/qq_37466121/article/details/87719044
ROC曲线的含义以及画法
https://blog.csdn.net/m0_48520385/article/details/118636338
课程资料包括糖尿病预测模型代码(SVM、随机森林、逻辑回归、K近邻、梯度提升模型,基于python)、论文的NHANES数据、复现论文、讲义。关注公众号“熊大学习社”,回复“t221201”,可获得资料链接。
另外,我们提供专业的SCI论文指导,服务合作请加vx,见主页下方。
感谢您的学习,希望您有所收获。您的一键三连是我最大的动力。
更多的学习分享,关注B站熊大学习社。
