时间:2024-09-01 17:01:22 阅读:181

诺维医学科研官网:https://www.newboat.top 我们的课程比丁香园更香!
Bilibili:文章对应的讲解视频在此。熊大学习社 https://space.bilibili.com/475774512
微信公众号|Bilibili|抖音|小红书|知乎|全网同名:熊大学习社
医学资源网站,https://med.newboat.top/ ,内有医学离线数据库、数据提取、科研神器等高质量资料库
课程相关资料:
(1)课程资料包括[DAY1]SCI论文复现全部代码-基于R、PostgreSql/Navicat等软件、SQL常用命令与批处理脚本、讲义;[Day2]MIMIC IV常见数据提取代码-基于sql、数据清洗-基于R讲义;[Day3] 复现论文、复现代码、复现数据、学习资料、讲义[Day4]扩展学习资料和相关源码等。关注公众号“熊大学习社”,回复“mimic01”,获取全部4天MIMIC复现课程资料链接。
(2)医学公共数据数据库学习训练营已开班,论文指导精英班学员可获取推荐审稿人信息,欢迎咨询课程助理!
(3)关注熊大学习社。您的一键三连是我最大的动力。


Day1:一、MIMIC数据库零基础入门
(1)MIMIC数据库获取
(2)MIMIC数据库软件安装
(3)MIMIC数据表介绍、基础数据提取
Day2:二、MIMIC数据库数据提取与清洗
(1)物化视图安装与简介
(2)关键数据提取与实操
(3)数据清洗实操
Day3:三、MIMIC数据库SCI论文复现上(生存分析SCI论文研究思路复现)
(1)数据提取。数据提取的实操方法分享,4条原则。
(2)PSM倾向评分。
(3)生存分析。基于PSM样本前和样本后的数据,survfit非参数生存分析。
(4)Cox风险比例模型分析和森林图。
(5)RCS限制性立方样条图。包括直方图、RCS图、RCS+直方图、年龄分组RCS+直方图、基于ggrcs的方法。
(6)亚组分析和森林图。
Day4:四、MIMIC数据库SCI论文复现下(接上)
这次直播课程的特点:上手操作+撸代码,零基础到SCI复现,随时互动交流,快速开启你的医学研究。

网址:https://pubmed.ncbi.nlm.nih.gov/37560475/
文章在学习资料包
论文名称:Aspirin reduces the mortality risk of sepsis-associated acute kidney injury: an observational study using the MIMIC IV database(阿司匹林降低败血症相关急性肾损伤的死亡率:一项使用MIMIC IV数据库的观察性研究)
论文数据分析方法:基线分析+PSM倾向评分匹配+生存分析+Cox风险比例模型+亚组分析。
作为对应,我们的主题:阿司匹林及其与P2Y12受体拮抗剂和败血症重症患者的全因死亡率的关联性:基于MIMIC IV数据库
P2Y12受体拮抗剂是一类作用于血小板P2Y12受体,对二磷酸腺苷引起的血小板聚集起抑制作用的药物,临床上主要用于预防和治疗心血管疾病的血栓事件。
复现数据分析方法:数据提取+缺失值插补+基线分析+PSM倾向评分匹配+生存分析+Cox风险比例模型+RCS限制立方样条图+亚组分析。
(1) sepsis脓毒症
(2) 抗血小板用药:asipirin阿司匹林、clopidogrel氯吡格雷、ticagrelor替卡格雷
(3) 抗凝血用药:heparin肝素
(4) 身高和体重数据
(5) 凝血指标
(6) 血检数据
方法1:大文件用数据库SQL命令进行初步提取并导出为csv文件。
方法2:小文件直接从csv读取,在RStudio中读取。
方法3:指定的切片数据从csv.gz读取。
方法4:数据提取汇总到RStudio进行数据预处理。
用方法2
sepsis <- fread("derived/sepsis3.csv") %>%
left_join(fread("derived/icustay_detail.csv")) %>%
left_join(select(fread("hosp/admissions.csv"), hadm_id, deathtime)) %>%
distinct(subject_id, hadm_id, stay_id, .keep_all = TRUE) %>%
mutate(race = case_when(
grepl("BLACK", race) ~ "Black",
grepl("WHITE", race) ~ "White",
grepl("ASIAN", race) ~ "Asian",
grepl("HISPANIC", race) ~ "Hispanic",
grepl("OTHER", race) ~ "Other"
))
asipirin阿司匹林
先用方法1,读取数据库数据。新建一个schema。
CREATE SCHEMA proj_sepsis;
查询阿莫斯林aspirin的药方数据,并保存。
CREATE TABLE proj_sepsis.aspirin AS (
SELECT * FROM mimiciv_hosp.prescriptions WHERE LOWER(drug) like '%aspirin%'
)
数据导出为asipirin.csv,后面在RStudio要用。
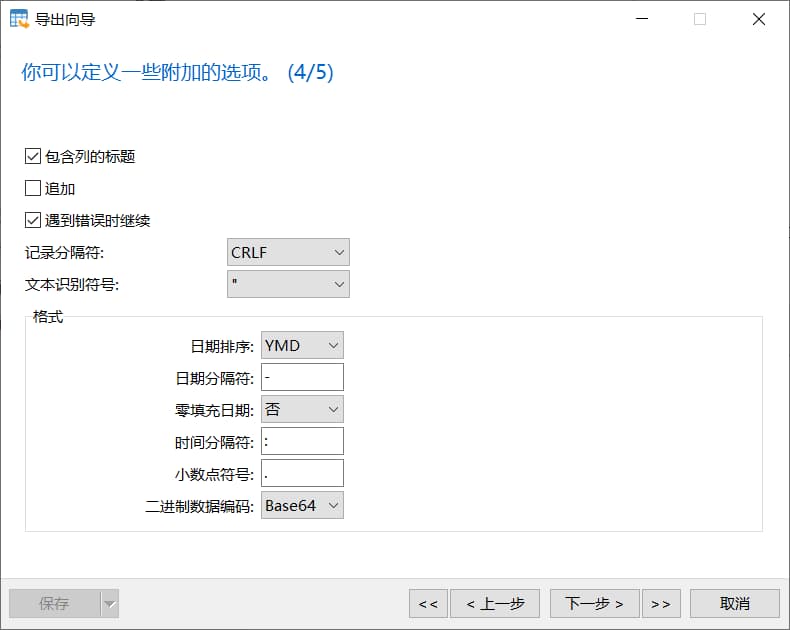
需要注意的是:日期排序
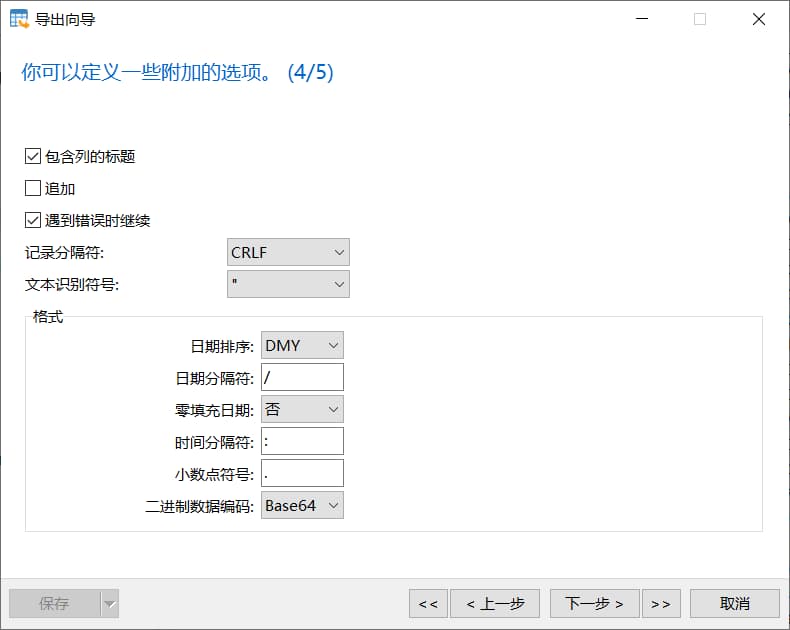
保存前:
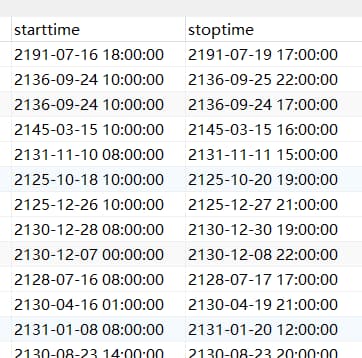
保存后:
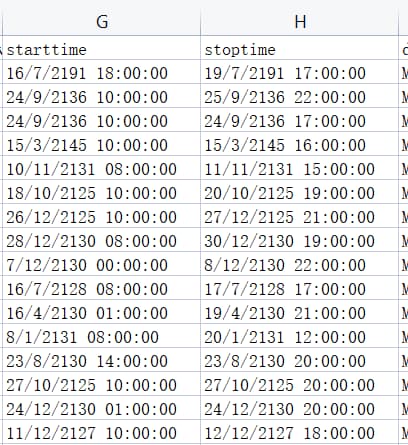
正确的做法:为保持一致,日期排序选YMD,日期分隔符为-。
然后用方法2,在RStudio中处理。
# 设置代码目录
setwd(code_path)
getwd()
aspirin_sorts <- read.csv("aspirin.csv")
aspirin_sorts$drug_extracted <- 'aspirin'
aspirin <- aspirin_sorts %>%
# 筛选出hadm_id在sepsis数据集中的记录
filter(hadm_id %in% sepsis$hadm_id)%>%
# 左连接合并sepsis数据集中的hadm_id和icu_intime列
left_join(select(sepsis, hadm_id, icu_intime)) %>%
group_by(hadm_id) %>%
filter(stoptime!='')%>%
# 筛选出stoptime大于等于icu_intime且stoptime大于starttime的记录
filter(stoptime >= icu_intime & stoptime > starttime) %>%
ungroup()%>%
# 将dose_val_rx列转换为数值型
mutate(dose_val_rx = as.numeric(dose_val_rx)) %>%
group_by(hadm_id) %>%
# 汇总计算每个hadm_id的dose_val_rx的最小值、最大值、starttime的最小值、stoptime的最大值
summarise(
dose_min = min(dose_val_rx, na.rm = TRUE),
dose_max = max(dose_val_rx, na.rm = TRUE),
starttime = min(starttime),
stoptime = max(stoptime)
)
# 计算处方持续时间(duration_pres)
aspirin <- aspirin %>%
mutate(
duration_pres = ifelse(
# 判断停止时间(stoptime)的小时是否大于等于8
lubridate::hour(stoptime) >= 8,
# 大于等于8小时,则取停止时间的天数减去开始时间(starttime)的天数
ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days"),
# 小于8小时,则取停止时间的天数减去开始时间的天数再减1
ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days") - 1
)
)
clopidogrel氯吡格雷
先用方法一,查询氯吡格雷clopidogrel的药方数据,并保存。
CREATE TABLE proj_sepsis.clopidogrel AS (
SELECT * FROM mimiciv_hosp.prescriptions WHERE LOWER(drug) like '%clopidogrel%'
)
后用方法二。
clopidogrel_sorts <- read.csv("clopidogrel.csv")
clopidogrel_sorts$drug_extracted <- 'clopidogrel'
clopidogrel <- clopidogrel_sorts %>%
# 筛选出hadm_id在sepsis数据集中的记录
filter(hadm_id %in% sepsis$hadm_id)%>%
# 左连接合并sepsis数据集中的hadm_id和icu_intime列
left_join(select(sepsis, hadm_id, icu_intime)) %>%
group_by(hadm_id) %>%
filter(stoptime!='')%>%
# 筛选出stoptime大于等于icu_intime且stoptime大于starttime的记录
filter(stoptime >= icu_intime & stoptime > starttime) %>%
ungroup()%>%
# 将dose_val_rx列转换为数值型
mutate(dose_val_rx = as.numeric(dose_val_rx)) %>%
group_by(hadm_id) %>%
# 汇总计算每个hadm_id的dose_val_rx的最小值、最大值、starttime的最小值、stoptime的最大值
summarise(
dose_min = min(dose_val_rx, na.rm = TRUE),
dose_max = max(dose_val_rx, na.rm = TRUE),
starttime = min(starttime),
stoptime = max(stoptime)
)
# 计算处方持续时间(duration_pres)
clopidogrel <- clopidogrel %>%
mutate(
duration_pres = ifelse(
# 判断停止时间(stoptime)的小时是否大于等于8
lubridate::hour(stoptime) >= 8,
# 大于等于8小时,则取停止时间的天数减去开始时间(starttime)的天数
ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days"),
# 小于8小时,则取停止时间的天数减去开始时间的天数再减1
ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days") - 1
)
)
ticagrelor替卡格雷
先用方法一,查询替卡格雷ticagrelor的药方数据,并保存。
CREATE TABLE proj_sepsis.ticagrelor AS (
SELECT * FROM mimiciv_hosp.prescriptions WHERE LOWER(drug) like '%ticagrelor%'
)
后用方法二。
ticagrelor_sorts <- read.csv("ticagrelor.csv")
ticagrelor_sorts$drug_extracted <- 'ticagrelor'
ticagrelor <- ticagrelor_sorts %>%
# 筛选出hadm_id在sepsis数据集中的记录
filter(hadm_id %in% sepsis$hadm_id)%>%
# 左连接合并sepsis数据集中的hadm_id和icu_intime列
left_join(select(sepsis, hadm_id, icu_intime)) %>%
group_by(hadm_id) %>%
filter(stoptime!='')%>%
# 筛选出stoptime大于等于icu_intime且stoptime大于starttime的记录
filter(stoptime >= icu_intime & stoptime > starttime) %>%
ungroup()%>%
# 将dose_val_rx列转换为数值型
mutate(dose_val_rx = as.numeric(dose_val_rx)) %>%
group_by(hadm_id) %>%
# 汇总计算每个hadm_id的dose_val_rx的最小值、最大值、starttime的最小值、stoptime的最大值
summarise(
dose_min = min(dose_val_rx, na.rm = TRUE),
dose_max = max(dose_val_rx, na.rm = TRUE),
starttime = min(starttime),
stoptime = max(stoptime)
)
# 计算处方持续时间(duration_pres)
ticagrelor <- ticagrelor %>%
mutate(
duration_pres = ifelse(
# 判断停止时间(stoptime)的小时是否大于等于8
lubridate::hour(stoptime) >= 8,
# 大于等于8小时,则取停止时间的天数减去开始时间(starttime)的天数
ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days"),
# 小于8小时,则取停止时间的天数减去开始时间的天数再减1
ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days") - 1
)
)%>%
# 列名更换,除了hadm_id,其他加上后缀
rename_at(vars(-c("hadm_id")), function(x) paste0(x, "_ticagrelor"))
heparin肝素
先用方法一,查询肝素heparin的药方数据,并保存。
CREATE TABLE proj_sepsis.heparin AS ( SELECT * FROM mimiciv_hosp.prescriptions WHERE LOWER(drug) like '%heparin%' AND LOWER(drug) not like '%flush%' )
后用方法二。
heparin_sorts <- read.csv("heparin.csv") heparin_sorts$drug_extracted <- 'heparin' heparin <- heparin_sorts %>% # 筛选出hadm_id在sepsis数据集中的记录 filter(hadm_id %in% sepsis$hadm_id)%>% # 左连接合并sepsis数据集中的hadm_id和icu_intime列 left_join(select(sepsis, hadm_id, icu_intime)) %>% group_by(hadm_id) %>% filter(stoptime!='')%>% # 筛选出stoptime大于等于icu_intime且stoptime大于starttime的记录 filter(stoptime >= icu_intime & stoptime > starttime) %>% ungroup()%>% # 将dose_val_rx列转换为数值型 mutate(dose_val_rx = as.numeric(dose_val_rx)) %>% group_by(hadm_id) %>% # 汇总计算每个hadm_id的dose_val_rx的最小值、最大值、starttime的最小值、stoptime的最大值 summarise( dose_min = min(dose_val_rx, na.rm = TRUE), dose_max = max(dose_val_rx, na.rm = TRUE), starttime = min(starttime), stoptime = max(stoptime) ) # 计算处方持续时间(duration_pres) heparin <- heparin %>% mutate( duration_pres = ifelse( # 判断停止时间(stoptime)的小时是否大于等于8 lubridate::hour(stoptime) >= 8, # 大于等于8小时,则取停止时间的天数减去开始时间(starttime)的天数 ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days"), # 小于8小时,则取停止时间的天数减去开始时间的天数再减1 ceiling_date(as.Date(stoptime), unit = "days") - floor_date(as.Date(starttime), unit = "days") - 1 ) )%>% # 列名更换,除了hadm_id,其他加上后缀 rename_at(vars(-c("hadm_id")), function(x) paste0(x, "_heparin"))## 2.1 抗血小板药方--------- antiplatelet <- full_join(aspirin, clopidogrel, by = "hadm_id", suffix = c("_aspirin", "_clopidogrel")) %>% full_join(ticagrelor, by = "hadm_id") ## 2.2 抗血小板药方的脓毒症患者---------- target_patient <- sepsis %>% left_join(antiplatelet) %>% mutate( aspirin = ifelse(hadm_id %in% aspirin$hadm_id, 1, 0), clopidogrel = ifelse(hadm_id %in% clopidogrel$hadm_id, 1, 0), ticagrelor = ifelse(hadm_id %in% ticagrelor$hadm_id, 1, 0), antiplatelet_sorts = aspirin + clopidogrel + ticagrelor ) %>% tidylog::filter(aspirin == 1) %>% tidylog::filter(antiplatelet_sorts != 3) %>% mutate(group = ifelse(antiplatelet_sorts == 1, 0, 1)) # 创建目标患者数据集(target_patient) target_patient <- sepsis %>% left_join(antiplatelet) %>% mutate( # 判断hadm_id是否在aspirin数据集中,是则为1,否则为0 aspirin = ifelse(hadm_id %in% aspirin$hadm_id, 1, 0), # 判断hadm_id是否在clopidogrel数据集中,是则为1,否则为0 clopidogrel = ifelse(hadm_id %in% clopidogrel$hadm_id, 1, 0), # 判断hadm_id是否在ticagrelor数据集中,是则为1,否则为0 ticagrelor = ifelse(hadm_id %in% ticagrelor$hadm_id, 1, 0), # 新增一列,抗血小板药物种类总数 antiplatelet_sorts = aspirin + clopidogrel + ticagrelor ) # 保留aspirin为1的记录 target_patient <- target_patient %>% tidylog::filter(aspirin == 1) # 保留antiplatelet_sorts不等于3的记录 # 排除3种药物都服用的数据 target_patient <- target_patient %>% tidylog::filter(antiplatelet_sorts != 3) # 分组变量,等于1的为0,否则为1 # 分成2组:一组为只服用aspirin,一组为服用aspirin以及另一种药(clopidogrel或ticagrelor) target_patient <- target_patient %>% mutate(group = ifelse(antiplatelet_sorts == 1, 0, 1))# 体重数据 weight <- fread("derived/first_day_weight.csv") %>% filter(stay_id %in% target_patient$stay_id) %>% select(stay_id, weight) # 身高数据 height <- fread("derived/first_day_height.csv") %>% filter(stay_id %in% target_patient$stay_id)# 糖尿病 diabetes_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(2313:2372, 16339:16692) # 高血压 hypertension_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(4650:4682, 24499:24521) # 肿瘤 neoplasm_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(1275:2332, 13534:15627) # 慢性阻塞性肺疾病 COPD_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(26410:26414) # 肿瘤手术 CA_surgery_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(c(99666, 99667, 109654, 109658, 109749) - 1) # 胃肠道 GI_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(c(5063, 5065, 5592, 5607, 5608, 53120, 53121, 53200, 53201, 53220, 53221, 53300, 53301, 53320, 53321, 53400, 53401, 53420, 53421, 27628, 26044, 26047, 26899, 26917, 26925, 26927, 26935, 26937, 26945, 26947, 26978, 26981, 26994) - 1) # 颅内出血 ICH_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(4851:4855, 24823:24866) # 静脉血栓栓塞 VTE_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(c(4740:4743, 4746, 7333:7337, 24627:24637, 5008:5025, 5038:5046, 25726:25841, 25925:25936) - 1) # 脑梗死 CI_icd <- fread("../mimic-iv-2.2/hosp/d_icd_diagnoses.csv.gz") %>% slice(c(4870, 4872, 24868:24983) - 1) # 患者确诊疾病信息 comorbidities <- fread("../mimic-iv-2.2/hosp/diagnoses_icd.csv.gz") %>% filter(hadm_id %in% target_patient$hadm_id) %>% mutate( # 糖尿病 co_diabetes = ifelse(icd_code %in% diabetes_icd$icd_code, 1, 0), # 高血压 co_hypertension = ifelse(icd_code %in% hypertension_icd$icd_code, 1, 0), # 肿瘤 co_neoplasm = ifelse(icd_code %in% neoplasm_icd$icd_code, 1, 0), # 慢性阻塞性肺疾病 co_COPD = ifelse(icd_code %in% COPD_icd$icd_code, 1, 0), # 肿瘤手术 co_CA_surgery = ifelse(icd_code %in% CA_surgery_icd$icd_code, 1, 0), # 胃肠道 co_GI = ifelse(icd_code %in% GI_icd$icd_code, 1, 0), # 颅内出血 co_ICH = ifelse(icd_code %in% ICH_icd$icd_code, 1, 0), # 胃肠道相关或颅内出血 co_bleeding = if_else(co_GI == 1 | co_ICH == 1, 1, 0), # 静脉血栓栓塞 co_VTE = ifelse(icd_code %in% VTE_icd$icd_code, 1, 0), # 脑梗死 co_CI = ifelse(icd_code %in% CI_icd$icd_code, 1, 0) ) # 疾病如胃肠道,需诊断3次以上,才能确诊;不到3次的数据排除 comorbidities <- comorbidities %>% tidylog::filter(!(icd_code %in% GI_icd$icd_code & seq_num < 3)) %>% tidylog::filter(!(icd_code %in% ICH_icd$icd_code & seq_num < 3)) %>% tidylog::filter(!(icd_code %in% VTE_icd$icd_code & seq_num < 3)) %>% tidylog::filter(!(icd_code %in% CI_icd$icd_code & seq_num < 3)) # 按hadm_id分组,并求各列最大值 comorbidities <- comorbidities %>% group_by(hadm_id) %>% summarise( co_diabetes = max(co_diabetes), co_hypertension = max(co_hypertension), co_neoplasm = max(co_neoplasm), co_COPD = max(co_COPD), co_CA_surgery = max(co_CA_surgery), co_GI = max(co_GI), co_ICH = max(co_ICH), co_bleeding = max(co_bleeding), co_VTE = max(co_VTE), co_CI = max(co_CI) )# 凝血数据 coagulation_baseline <- fread("derived/coagulation.csv") %>% mutate( starttime = floor_date(charttime, unit = "hour"), endtime = ceiling_date(charttime, unit = "hour") ) %>% select(-specimen_id) %>% rename("charttime_coagulation" = "charttime") %>% filter(hadm_id %in% target_patient$hadm_id) %>% group_by(hadm_id) %>% # 按照charttime_coagulation升序排列数据 arrange(charttime_coagulation) %>% # 保留每个hadm_id的第一条记录 slice(1) %>% # 选择以"id"结尾的列以及d_dimer到ptt列 select(ends_with("id"), d_dimer:ptt)# 血细胞计数 blood_count_baseline <- fread("derived/complete_blood_count.csv") %>% mutate( starttime = floor_date(charttime, unit = "hour"), endtime = ceiling_date(charttime, unit = "hour") ) %>% select(-specimen_id) %>% rename("charttime_blood" = "charttime") %>% filter(hadm_id %in% target_patient$hadm_id) %>% group_by(hadm_id) %>% # 按照charttime_blood升序排列数据 arrange(charttime_blood) %>% # 保留每个hadm_id的第一条记录 slice(1) %>% # 选择以"id"结尾的列以及d_dimer到ptt列 select(ends_with("id"), hematocrit:wbc)## 3.1 数据合并------ data_raw <- target_patient %>% left_join(heparin) %>% left_join(height) %>% left_join(weight) %>% left_join(comorbidities) %>% left_join(coagulation_baseline) %>% left_join(blood_count_baseline)
## 3.2 数据纳排------ data_raw <- data_raw %>% tidylog::filter(admission_age >= 18) %>% tidylog::filter(los_icu > 2) %>% tidylog::filter(first_icu_stay == "t") %>% tidylog::filter(duration_pres_aspirin >= 3 | is.na(duration_pres_aspirin) | duration_pres_clopidogrel >= 3 | is.na(duration_pres_clopidogrel) | duration_pres_ticagrelor >= 3 | is.na(duration_pres_ticagrelor)) %>% ungroup() %>% tidylog::drop_na(co_diabetes:co_CA_surgery)
## 3.3 数据预处理------ data_raw <- data_raw %>% mutate( # 生存时间 surv_time = as.double(as.period(ymd_hms(deathtime) - ymd_hms(sofa_time)), units = "days"), # 分为2组:是否超过30天 surv_30 = case_when( surv_time >= 30 ~ 30, surv_time == NA ~ 30, TRUE ~ surv_time ), # 分为2组:是否超过90天 surv_90 = case_when( surv_time >= 90 ~ 90, surv_time == NA ~ 90, TRUE ~ surv_time ), # 生存为0,死亡为1 status_30 = ifelse(surv_30 == 30, 0, 1), status_90 = ifelse(surv_90 == 90, 0, 1) ) %>% # 空值处理 replace_na( list( surv_30 = 30, surv_90 = 90, status_30 = 0, status_90 = 0 ) )
对缺失值进行估算填充的目的不是为了准确的估算缺失值真实的值,而是为了预测缺失值所服从的分布
【参考文章】R语言处理缺失值的多重插补技术-mice包
https://zhuanlan.zhihu.com/p/610103488
## 3.4 多重插补multiple imputation------ baseline <- data_raw %>% select( # 列名重命名 Age = admission_age, Weight = weight, Height = height, Gender = gender, Race = race, Hypertension = co_hypertension, Diabetes = co_diabetes, Neoplasm = co_neoplasm, COPD = co_COPD, `History of CAS` = co_CA_surgery, INR = inr, PT = pt ) # 图形探究缺失值 baseline %>% VIM::aggr(prop=T, numbers=F, cex.label = 1.2, cex.aixs = 2, gap = 3, labels = names(.), oma = c(8, 6, 2, 3)) # 查看缺失值数量和比例 baseline %>% naniar::miss_var_summary() # 缺失值的可视化 baseline %>% visdat::vis_miss() # 完全随机缺失检验 baseline %>% naniar::mcar_test() # 均值插补:weight/inr/pt data_simple_impute <- data_raw %>% replace_na( list( # 当na.rm设置为TRUE时,函数会自动忽略缺失值并进行计算 weight = median(.$weight, na.rm = T), inr = median(.$inr, na.rm = T), pt = median(.$pt, na.rm = T) ) ) # 随机插补:race/height imp <- data_simple_impute %>% select(admission_age, weight, height, gender, race, co_diabetes, co_hypertension, co_neoplasm, co_COPD, co_CA_surgery, inr, pt) %>% mutate(race = as.factor(race)) %>% # 随机森林插补rf,1次插补 mice(method = "rf", m = 1, seed = 2024) # 查看插补变量和插补方法 imp$method # 生成插补数据 data_imputed <- complete(imp) %>% select(height, race) %>% rename_at(c(names(.)), function(x) paste0(x, "_imputed")) %>% as_tibble() # 零值插补:duration_pres_heparin # 得到最终数据 final_data <- data_simple_impute %>% bind_cols(data_imputed) %>% mutate( race = as.factor(race), gender = as.factor(gender), race = factor(race, levels = c("White", "Black", "Asian", "Hispanic", "Others")), co_diabetes = as.factor(co_diabetes), co_hypertension = as.factor(co_hypertension), co_neoplasm = as.factor(co_neoplasm), co_COPD = as.factor(co_COPD), co_CA_surgery = as.factor(co_CA_surgery), co_VTE = as.factor(co_VTE), co_CI = as.factor(co_CI), co_GI = as.factor(co_GI), co_ICH = as.factor(co_ICH), co_bleeding = as.factor(co_bleeding), group = as.factor(group) ) %>% replace_na( list( duration_pres_aspirin = 0, duration_pres_clopidogrel = 0, duration_pres_ticagrelor = 0, duration_pres_heparin = 0 ) )在因果推断中,PSM指的是倾向评分匹配(Propensity Score Matching)。它是一种常见的非实验性因果推断方法,用于减少观察数据集中存在的选择偏差或混杂变量的影响。PSM的主要目标是通过建立一个“倾向得分”来估计处理组和对照组之间的因果效应。
PSM的一般步骤包括:
估计每个个体进入处理组的概率,即倾向得分。
根据倾向得分使用一种匹配算法来创建处理组和对照组之间的匹配对。
在匹配后,可以比较处理组和对照组之间的平均效应差异,从而估计因果效应。
尽管PSM在某些情况下可以帮助控制潜在的混杂变量,但它仍然依赖于某些假设,例如倾向得分模型的正确规范和匹配过程的有效性。因此,在使用PSM时需要小心,并结合其他方法进行敏感性分析以确保结果的可靠性。
匹配方法:最近邻配比法(nearest neighbor matching)、卡钳匹配/半径匹配/卡尺匹配(caliper matching/radius matching)、马氏矩阵配比法(Mahalanobis metric matching)、核匹配(kernel matching)。
## 4.1 生成PSM样本--------- set.seed(2024) # 选取数据 data_tidy <- final_data %>% select(hadm_id, admission_age, gender, height_imputed, weight, starts_with("sofa"), race_imputed, starts_with("co_"), duration_pres_heparin, inr, pt, los_icu, los_hosp = los_hospital, surv_30, status_30, surv_90, status_90, group, -sofa_time) # PSM psm <- data_tidy %>% matchit( formula = group ~ admission_age + gender + weight + height_imputed + sofa_score + race_imputed + co_diabetes + co_hypertension + co_neoplasm + co_COPD + co_CA_surgery + duration_pres_heparin + inr + pt, distance = "glm", method = "nearest", ratio = 1, caliper = 0.05 ) summary(psm) # 匹配后样本数据 data_psm <- match.data(psm)%>% select(-c(distance:subclass)) %>% as_tibble() %>% mutate( group = as.factor(group) ) # 保存PSM后样本数据 write_csv(data_psm, file = "data_psm.csv") # 绘制倾向评分的分布情况 plot(psm, type = "jitter", interactive = FALSE) # 查看指定变量的分布 plot(psm, type = "qq", interactive = FALSE, which.xs = c("admission_age", "sofa_score")) # qq分布 plot(psm, type = "hist") # Standardized Mean Differences,标准化平均差 # 差异小于0.1,则表示匹配效果较好 plot(summary(psm))## 4.2 基线分析:CreateTableOne------- colnames(data_psm) str(data_psm) # 变量集合 myVars <- c("gender", "admission_age", "height_imputed", "weight", "sofa_score", "race_imputed", "co_diabetes", "co_hypertension", "co_neoplasm", "co_COPD", "co_CA_surgery", "co_GI", "co_ICH", "co_bleeding", "co_VTE", "co_CI", "duration_pres_heparin", "inr", "pt", "los_icu", "los_hosp", "surv_30", "status_30", "surv_90", "status_90") # 分类变量 catVars <- c("gender", "race_imputed", "co_diabetes", "co_hypertension", "co_neoplasm", "co_COPD", "co_CA_surgery", "co_GI", "co_ICH", "co_bleeding", "co_VTE", "co_CI", "status_30", "status_90") # 连续变量正态性检验 # p>0.05才符合正态分布 shapiro.test(data_psm$admission_age) # 非正态分布变量集合,有的话写上 nonvar <- c("") # PSM前基线表 table1 <- CreateTableOne(vars = myVars, # 全部变量,自变量+协变量 strata = "group", # 结局变量 data = data_tidy, # 数据 factorVars = catVars, # 分别变量 addOverall = TRUE) # 显示总体样本情况 #将生成的table1存成csv格式 print(table1, showAllLevels = TRUE, # 显示所有变量 nonnormal = nonvar, # 不符合正态性分布,基线表汇报中位数+四分位数 smd = F) |> write.csv(file = "Talbe1 baseline.csv") # PSM后基线表 table2 <- CreateTableOne(vars = myVars, # 全部变量,自变量+协变量 strata = "group", # 结局变量 data = data_psm, # 数据 factorVars = catVars, # 分别变量 addOverall = TRUE) # 显示总体样本情况 #将生成的table1存成csv格式 print(table2, showAllLevels = TRUE, # 显示所有变量 nonnormal = nonvar, # 不符合正态性分布,基线表汇报中位数+四分位数 smd = F) |> write.csv(file = "Talbe1 baseline_psm.csv")## 4.3 【自学】连续变量检验------- # 主要是正态分布检验 normality # 连续变量,即数字类型的列名 numeric_var <- data_psm %>% select_if(is.numeric) %>% select(-c(hadm_id, status_30, status_90, surv_30, surv_90)) %>% names() ### 4.3.1 Anderson–Darling AD检验----------- ad.test.multi <- function(x) { ad.test(x) %>% broom::tidy() } # AD检验结果 normality <- map_df(data_psm[numeric_var], ad.test.multi) %>% # 增加一行,变量名 bind_cols(variable = numeric_var) %>% # 选取指定列并改名 select(variable, statistic, p_norm='p.value')### 4.3.2 方差检验variance test---------------- variance <- map_df(data_psm[numeric_var], function(x) {levene_test(data_psm, x ~ data_psm$group)}) %>% select(p_vari = p) %>% cbind(variable = numeric_var)pivot_longer,横表变竖表
【参考文章】R语言数据reshape:dplyr包函数pivot_longer https://blog.csdn.net/weixin_43168119/article/details/135899807
### 4.3.3 t检验 t test------------ t_test_res <- data_psm %>% # 从PSM样本中选择数值变量和分组变量 select(all_of(numeric_var), group) %>% # 横表变竖表 pivot_longer( cols = all_of(numeric_var), names_to = "variable" ) %>% group_by(variable) %>% # t检验,比较不同组之间的差异 t_test(value ~ group, var.equal = TRUE, paired = TRUE) %>% # 选取指定列及改名 select(variable, p_t_test = p)
### 4.3.4 wilcox秩和检验 wilcox test--------- wilcox_test_res <- data_psm %>% # 从PSM样本中选择数值变量和分组变量 select(all_of(numeric_var), group) %>% # 横表变竖表 pivot_longer( cols = all_of(numeric_var), names_to = "variable" ) %>% group_by(variable) %>% # t检验,比较不同组之间的差异 wilcox_test(value ~ group, paired = TRUE) %>% # 选取指定列及改名 select(variable, p_wilcox_test = p)
### 4.3.5 连续变量检验汇总------------ p_numb <- normality %>% left_join(variance, by = "variable") %>% left_join(t_test_res, by = "variable") %>% left_join(wilcox_test_res, by = "variable") %>% group_by(variable) %>% mutate(p_final = if_else(p_norm >= 0.05 & p_vari >= 0.05, p_t_test, p_wilcox_test)) %>% add_significance(p.col = "p_final")
### 4.4.1 卡方独立性检验 chisq test ----------- p_chisq <- data_psm %>% select(all_of(nominal_var), group) %>% # # 横表变竖表 pivot_longer( cols = -group, names_to = "variable" ) %>% group_by(variable) %>% # 卡方独立性检验 chisq test do(chisq_test(.$group, .$value)) %>% select(variable, p_chisq = p)
### 4.4.2 fisher检验------------ # 统计样本数据的频次和概率 freq <- freq_table(data_psm, group, all_of(nominal_var)) fisher_map <- function(x) { temp <- freq_table(data_psm, group, x) %>% # 删除prop列 select(-prop) %>% # 横表变竖表 pivot_wider( names_from = "group", values_from = "n" ) %>% # 空置设为0 replace(is.na(.), 0) p_fisher <- temp %>% select(all_of(unique(data_psm$group))) %>% fisher_test(simulate.p.value = TRUE) min_n <- temp %>% pivot_longer( cols = all_of(unique(data_psm$group)) ) %>% arrange(value) %>% slice(1) %>% select(value) %>% cbind(variable = x) cbind(p_fisher, min_n) } p_fisher <- map_df(all_of(nominal_var), fisher_map) %>% select(p_fisher = p, min_n = value, variable)### 4.4.3 分类变量检验汇总------------ p_norm <- tibble(variable = all_of(nominal_var)) %>% left_join(p_chisq, by = "variable") %>% left_join(p_fisher, by = "variable") %>% group_by(variable) %>% mutate(p_final = ifelse(nrow(data_psm) > 40 & min_n >= 5, p_chisq, p_fisher))
### 4.4.4 P值汇总--------- # 连续变量p值 p_numb # 分类变量p值 p_norm # 汇总 p_value <- bind_rows( p_numb %>% select(variable, p_final), p_norm %>% select(variable, p_final) )
theme_gtsummary_language("en", big.mark = "") psm_summary <- data_psm %>% # 基线分析 tbl_summary( by = group, statistic = list( all_continuous2() ~ c("{mean} u00B1 {sd}", "{median} ({p25}, {p75})", "{p_miss}"), all_categorical() ~ "{n} ({p}%)" ), digits = list( all_continuous() ~ 1, all_categorical() ~ 1 ) ) %>% # 添加p值 add_p() psm_summary %>% as_tibble() %>% rename("variable" = `**Characteristic**`) %>% left_join(p_value) %>% mutate(p_final = ifelse(p_final >= 0.001, round(p_final, 3), "< 0.001")) %>% write_csv(file = "psm_summary.csv")(1)学习资料补充,详见学习资料包。更多论文数据提取和分析的代码,详见学习资料包的Day4扩展资料。
中文翻译的d_icd_cn文件,csv直接导入。
(2)课程资料包括[DAY1]SCI论文复现全部代码-基于R、PostgreSql/Navicat等软件、SQL常用命令与批处理脚本、讲义;[Day2]MIMIC IV常见数据提取代码-基于sql、数据清洗-基于R讲义;[Day3] 复现论文、复现代码、复现数据、学习资料、讲义[Day4]扩展学习资料和相关源码等。关注公众号“熊大学习社”,回复“mimic01”,获取全部4天MIMIC复现课程资料链接。
(3)医学公共数据数据库学习训练营已开班,论文指导精英班学员可获取推荐审稿人信息,欢迎咨询课程助理!

(4)数据提取和数据分析定制,具体扫码咨询课程助理。